
(Introduction) With a large amount of genetic data available, computing power is needed to analyze and identify important genetic markers. Bioinformatics uses computer programs to analyze biological data. I had a chance to learn more about bioinformatics when I attended the Waksman Institute Summer Experience program in 2021. I also took a Python online course, the University of Michigan’s “Python for Everybody” course on Coursera. These are my notes.
Python is a popular general-purpose computer programming language that is commonly used in many industries, including in the biological field. It is easy to learn and can be used for text manipulation, writing to files, data analysis, visualizations, etc. Python is an interpreted language. The Python code is written in human understandable text, usually stored in a file with an extension “.py”. When the code is run, it is converted to bytecode, an intermediary language. Then the bytecode is executed by a Python interpreter program. This makes it easier to debug the code.

Python can be downloaded from the python.org download site and installed on your computer. This installs the Python interpreter program and associated tools that are needed to run Python code. The instructions to download are on this page. There are useful tutorials available at learnpython.org.
An Integrated Development Environment (IDE) is used to efficiently write programs in Python, though Python code can also be run via the command line. I use JetBrains PyCharm Community Edition which is free to download here.
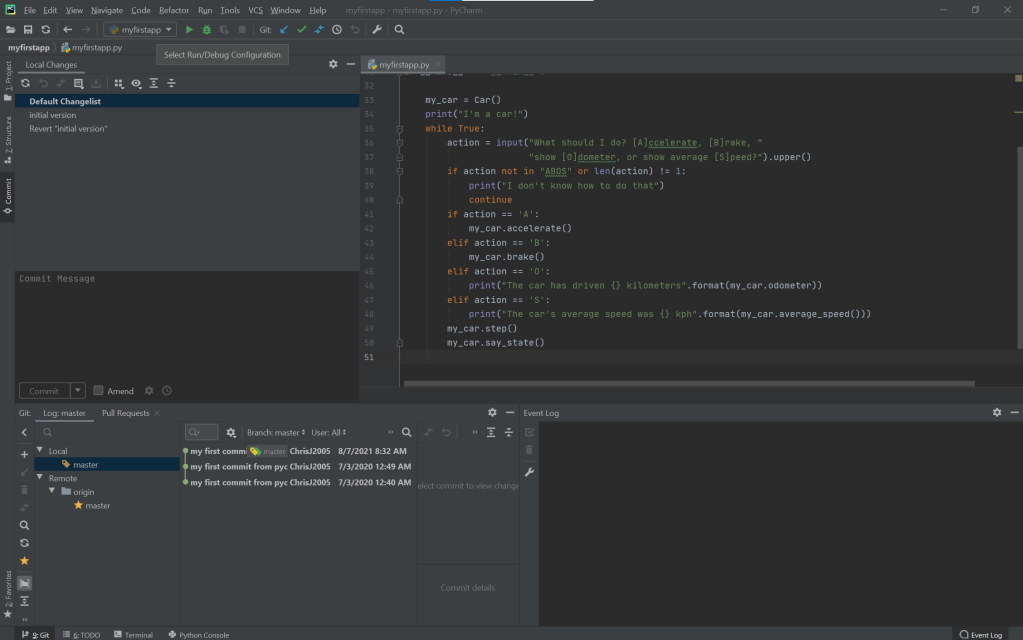
Here’s an example of a simple Python code that displays the text between the brackets. The output of the code is displayed too.
# This is the comment - my first python code!
print("Hello! This is Chris!")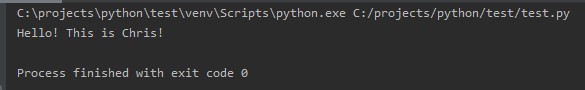
I will be adding more code snippets with notes in subsequent posts! Stay tuned!


Hey, great read! I particularly enjoyed your in-depth discussion of Python’s install, since it was something I hadn’t really thought of before. Being a fellow tech blogger myself, I also really appreciate how organized and well-formatted everything was – it definitely made the content much more digestible overall. Keep up the awesome work, I’ll be sure to stay tuned to future posts!
LikeLike